Nucleic acids, like DNA and RNA, are made of small organic molecules, called nucleotides. Four different nucleotides exist for each nucleic acid (A, G, C, T for DNA and A, C, G, U for RNA). Put three nucleotides together and you have a codon (fig.1).
In a way, codons are the bridge between the DNA molecule and amino acids (the building blocks of proteins); the genetic code tells the cell where the bridge should lead to, that is, which triplet codon corresponds to which amino acid. The codon GAC, for instance, originates the amino acid aspartate, whereas the codon GAG translates into glutamine.

There are 64 different codons – the number of possible arrangements of 4 nucleotides in sequences of three – but only 20 amino acids, so some amino acids can be encoded by more than one codon (fig.2). What do these synonymous codons do in the cell, if their function is redundant? Researchers at the University of Cambridge tried to answer this question. They figured that if they reduced the number of codons in the genome of an organism, they would know whether the deleted triplets are necessary for the cell’s normal cell functioning.

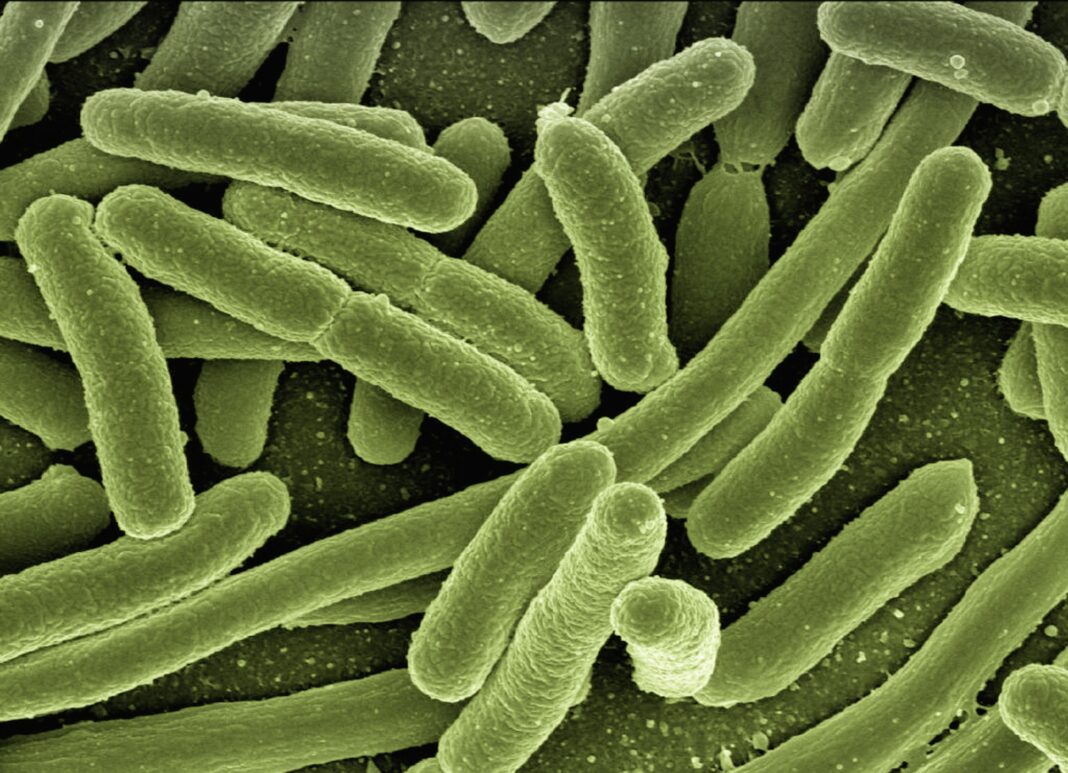
The first step was to build a synthetic genome, one where some redundant codons were replaced by their synonymous. The E. coli was the chosen organism to test whether these changes in DNA redundancies would cause damages to the cell’s machinery. Thus, the Syn61 was created, an E. coli with 61 codons in its genome, instead of the regular 64.
Apart from an initial delay in the Syn61’s growth, compared to the regular E. coli, the researchers report that the organism survives with normal cell functioning. One benefit of such discovery has to do with the study of viral infections: if the bacteria’s genome is different, it will be more difficult for viruses to invade them and replicate themselves within the organism. The discovery can also be important in the synthesis of new drugs with E. coli.
Article: Fredens, J., Wang, K., de la Torre, D., Funke, L., Robertson, W., Christova, Y., Chia, T., Schmied, W., Dunkelmann, D., Beránek, V., Uttamapinant, C., Llamazares, A., Elliott, T. and Chin, J. (2019). Total synthesis of Escherichia coli with a recoded genome. Nature. [CLOSED ACCESS]